Entry Version:
Citation:
Pancreapedia: Exocrine Pancreas Knowledge Base, DOI: 10.3998/panc.2019.02
| Attachment | Size |
|---|---|
| 380.31 KB | |
| 1.89 MB |
1. Overview (General)
α-Amylase (1,4-α-D-glucan-glucanohydrolase, EC 3.2.1.1) is the primary digestive enzyme acting on starch or glycogen and is present in plants, animals, bacteria and fungi. Starch from plants is a high molecular weight polymer of glucose. It is made up of amylose, a straight-chain α-1,4 linked polymer of about 105units and amylopectin, a branched chain polymer with α-1,4 linked glucose with branch points made up of α-1,6 linkages that contains in total about 106 units of glucose. Glycogen from animals is similar to amylopectin in structure but is smaller. α-Amylase cleaves the α-1,4 linkage when it is not next to a branch point or terminal glucose residue. Thus its products are maltose (2 glucose residues), maltotriose (3 glucose residues) and α-limit dextrins (5-6 residues which contain a branch point). In vertebrates, these are further cleaved to monosaccharides by the intestinal brush border enzymes isomaltase and maltase (glucoamylase) which hydrolyze the α-1,6 and α-1,4 bonds respectively (3). Studies show that hydrolysis and not monosaccharide absorption is the rate limiting step in complex carbohydrate absorption in humans (25).
In animals α-amylase occurs in pancreas, parotid, liver, serum, urine and occasionally in smaller amounts in other tissues or tumors; the major salivary and pancreatic amylase proteins are very similar (56). Salivary amylase initiates carbohydrate digestion in the mouth and pancreatic amylase is the main enzyme for luminal digestion of carbohydrate in the small intestine. Human pancreatic α-amylase is synthesized as a protein of 57 KDa for which the cDNA predicts a protein of 512 amino acids (66). This includes a signal sequence; amylase isolated from human pancreatic juice has 496 amino acids (20). In various species the reported molecular weight for amylase is 50-57 kDa and consists of a single chain protein with one carbohydrate (some species have an isoform with none) and an isoelectric point of 7.1. (47). Human pancreatic juice amylase has no sugar groups and exists as two isoforms of pI 7.2 and 6.6 termed HPA I and HPA II (20). Five disulfide bridges have been described in porcine pancreatic amylase (44) and most species have one or two free sulphydryl groups.
Salivary amylase is coded for by the AMY1 gene and pancreatic amylase by AMY2; a third form present in some tumors is termed AMY2B (41, 51, 68, 69). The evolution of the amylase gene family has been traced with a retroviral insertion into the promotor of a pancreatic amylase gene diverting it to become a salivary gene (38). Amylase is a multigene family with multiple genes and pseudogenes on chromosome 1 in humans and chromosome 3 in mice (24, 57). While most inbred mouse strains express a single species of amylase, some strains (A/J, CE/J) and most wild mice have multiple forms (6, 60). Interestingly, individuals from agricultural societies that consume high starch diets have a higher copy number of salivary amylase genes (45) and a high copy number leads to more protein and increased perception of oral starch (32, 50).
The structure of human amylase protein is known and contains three domains termed A, B, and C starting at the amino terminal (9). The active site is located the A domain; calcium binds to the B domain and may stabilize the active site (10). The C domain is a globular domain of unknown function. Some amylases including human pancreatic amylase are allosterically activated by chloride which modulates the pH optima and the maximal activity (15, 35).The active center of amylase contains 5 subsites which bind different glucose residues in the substrate (52). Current understanding of the enzymatic mechanism for glycosidases in reviewed in (63).
Kinetic evidence supports additional carbohydrate binding sites (2). Some of these are surface binding sites which help the amylase bind to starch granules (70). The rate of starch digestion also depends on the structure of the starch which has been classified as rapidly digested, slowly digested, and resistant starch. Digestion of starch can also be affected by cooking which can break down starch granules to soluble starch (17). Studies of the enzymatic mechanism have also benefited from the existence of a large family of plant α-amylase inhibitory proteins (7, 58, 62). The best studied of these are from beans and the structure of the enzyme-inhibitor complex has been determined (49). More potent inhibitors have been derived from pseudooligosaccarides of which one, the pseudotetrasacharide, acarbose has been approved for clinical use. The crystal structure of pig amylase complexed with acarbose has been reported (21). These pseudooligosaccaride inhibitors, in contrast to the proteinaceous inhibitors, block all glucosidases including intestinal brush border enzymes as well as amylase (53).
Multiple pancreatic enzymes including amylase are found absorbed to the surface of the duodenal mucosa although they could be washed off with high salt (3). Amylase, however, was shown to specifically bind to N-linked oligosaccharides of brush border glycoproteins and that binding enhanced amylase activity (33). Specific glycoproteins binding amylase have been identified as sucrase-isomaltase (SI) and SGLT1 (4). This binding enhanced the activity of SI as well as that of amylase but inhibited the activity of SGLT1. The authors proposed that this enhances digestion but prevents too rapid absorption of glucose. After binding to brush border proteins amylase is internalized into enterocytes restoring glucose uptake (16).
2. Pancreatic Function
Amylase Localization, Regulation and Deficiency
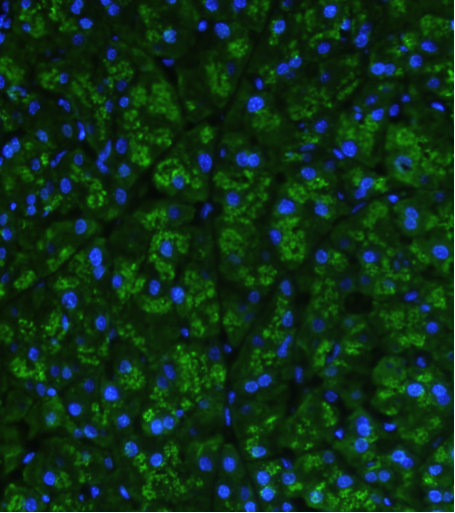
Pancreatic amylase is similar to other secretory proteins and is synthesized in the rough endoplasmic reticulum. It then moves through the Golgi apparatus and is stored in secretory granules until the time it undergoes regulated secretion. Immunostaining reveals its concentration in the granules (Figure 1). As with other pancreatic enzymes, the amount of amylase in the pancreas is regulated by the amount of dietary substrate (8, 22, 55). Rodents which eat a carbohydrate rich diet synthesize more amylase which is the most abundant pancreatic digestive enzyme in these species. This effect occurs at the transcriptional level and the effect is mediated in large part by insulin. With diabetes the pancreatic amylase falls, but can recover following insulin treatment (30, 61). This is especially prominent in rats where in experimental diabetes pancreatic amylase falls to only a few percent of normal. A 30 base pair insulin responsive element was identified in the mouse Amy2-2 gene (28). The fact that mice express more than one functional Amy2 gene explains why total pancreatic amylase does not fall as much in diabetes as it does in rats (18).
Figure 1: Immunoflourescent localization of amylase in mouse pancreas (green). Nuclei are stained blue with DAPI.
Healthy volunteers are able to absorb nearly 100% of a starch load. However, in chronic pancreatitis with loss of functioning pancreatic tissue, only about 90% is digested and absorbed; with complete inhibition of amylase this is reduced to 80-85% (31). In the latter case up to 20% of carbohydrate reaches the ileum and the rise of plasma GIP and C peptide from proinsulin is blocked. Patients with isolated amylase deficiency have been reported with carbohydrate malabsorption and its resulting symptoms (37, 54). The patients evaluated have intact mRNA but no amylase protein. This is similar to physiological pancreatic amylase deficiency at birth where lack of symptoms is in part due to low dietary starch requiring amylase and in part the action of salivary amylase. Amylase deficiency may be underreported due to mild symptoms.
Recently, the amount of pancreatic amylase mRNA and amylase activity in the jejunum of carbohydrate sensitive rats was shown to be related to dietary glucose oxidation and visceral fat accumulation (5)
Serum Amylase as a Measure of Pancreatic Function in Pancreatitis
Determination of serum amylase has long been a mainstay in the diagnosis of acute pancreatitis (1, 26). Serum amylase in normal individuals is about half salivary and half pancreatic form and each exists in several isoforms all of which contribute to total amylase. Because of the existence of different assays and standards, the cutoff for acute pancreatitis is usually expressed related to the range of normal for the particular clinical laboratory. Levels greater than three times the upper limit of normal (ULN) are usually taken as indicative of pancreatitis. However, the diagnostic specificity is only 70-90% depending on the population studied and the gold standard used (13, 64). A higher specificity is obtained when a higher cutoff point is used such as 5 or even 9 times the ULN, but at the cost of lower sensitivity. Utilizing assays to inhibit salivary amylase with wheat germ inhibitor or monoclonal antibodies (59) or removing it with immobilized monoclonal antibody (40) increase specificity somewhat and helps eliminate a contribution from tumors or other organs which primarily produce salivary amylase, but these tests are not always available. The combination of serum lipase with amylase is generally available and helps when serum amylase levels have already peaked and declined. A rise in serum amylase can also be due to macroamylassemia where amylase is bound to plasma protein (usually IgA or IgM) but this only occurs in only 1-3 % of normal humans. Amylase is also elevated in a variety of surgical, traumatic and neoplastic diseases (13, 46, 64). Renal disease can reduce amylase filtration and enhance plasma levels, but this increase is usually small as glomerular filtration is only a small portion of amylase clearance (46). A chronic increase in multiple serum pancreatic enzymes including amylase in otherwise healthy subjects without pancreatic disease was described by Gullo in 1996 and is now termed benign pancreatic hyperenzymemia or Gullo’s syndrome (23). Advanced imaging techniques may uncover a pancreatic abnormality in some of these patients.
It is also important to be aware that pancreatitis can occur without increased serum amylase especially with alcoholic pancreatitis or recurrent chronic pancreatitis (42). In some of these cases, lipase is elevated. Thus, while amylase is still one of the most useful tests in diagnosing acute pancreatitis, it should be carried out in conjunction with other tests (1) along with an understanding of amylase metabolism.
Amylase secretion as a measure of regulated pancreatic or parotid secretion
Measurement of amylase secretion has become widely adopted to measure regulated protein secretion by pancreas or parotid fragments, slices or isolated acini. Amylase has filled this role because it is easy to measure, sensitive, and does not require activation as do proteases. While for measurement of serum amylase in pancreatitis an absolute amount is desirable, most secretory measurements involve separately assaying the secretory product, usually in the incubation medium, and the tissue content and calculating the percent content released in a specified time. While earliest measurements assayed the reducing groups produced in the hydrolytic product, most often with dinitrosalicylic acid, studies over the last 50 years have more commonly used the solubilized products of substrate or a colored or flourescent enzymatic product produced in a coupled reaction. Examples of this approach are Remazolbrilliant blue starch (48), Procion Yellow labeled starch (27), amylopectin anthranilate (43), and Cibachronblue F3GA-amylose (29). A common commercially available substrate is the Phadebas reagent in which an unspecified blue dye is linked to starch (12). Most commonly these secretion studies are endpoint assays where tissue is incubated for a standard time and then aliquots of the medium are incubated with the starch substrate for a set time, centrifuged, and the product read in a spectrophotometer or flourometer (39, 67). Some of these assays have been adapted to continuous readout with a flow through system (14, 34). A second approach uses soluble colorless substrates that can be incubated in a plate reader with samples of the medium and lead to a colored or fluorescent product either directly or through a coupled reaction. One primary substrate is ethylidene-paranitrophenol-G7. This can be read directly or the glucose released determined by glucose oxidase (65). This type of assay has also been adapted to autoanalyzer technique. Recently both types of assays have been used to determine human duodenal amylase with high correlation (19).
Amylase Inhibitors in Clinical Medicine
Amylase inhibitors particularly Acarbose, a pseudotetrasacharide, have been used it the treatment of type 2 diabetes where they have moderate effects to reduce peak postprandial glucose and Hemoglobin A1c. Normally, they are used as a second line medication often in conjunction with metformin (36). Evidence also exists that Acarbose can reduce dumping syndrome after bariatric surgery (11). A host of natural plant products with amylase inhibitor activities have been proposed or studied as a treatment for diabetes but little conclusive evidence exists for their effectiveness.
3. Tools for the study of Amylase
a. Antibodies
Amylase antibodies are widely available but not always well tested. The most commonly used is from Sigma-Aldrich (cat # A8273) and is raised in rabbits injected with purified human salivary amylase. It is highly specific, reacts with both salivary and pancreatic amylase and in our hands works well for Western Blotting, immunofluorescence (Fig. 1) and immunohistochemistry in rodent tissue as well as human tissue. A number of companies sell anti-amylase antibodies raised in sheep, rat or mice (monoclonal) that would allow double labeling with other rabbit antibodies.
b. Assay kits and reagents
The most used amylase substrate for assays in research laboratories is the Phadebas reagent which is starch coupled to a blue dye. It was developed by Pharmacia Diagnostics and is now made by Magle Life Sciences and sold by Fisher Scientific. It can be used to measure amylase in plasma or serum as well as that secreted by pancreatic and parotid cells where it can be diluted to reduce cost. Alternative amylase assay kits are EnzChek Ultra Amylase Assay sold by ThermoFisher which uses bodipy labeled starch as substrate and Amylase Assay Kit from Sigma-Aldrich that uses ethylidine-paranitrophenol-G7 as substrate.
4. References
- Agarwal N, Pitchumoni CS, and Sivaprasad AV. Evaluating tests for acute pancreatitis. Am J Gastroenterol 85: 356-366, 1990. PMID: 2183590.
- Alkazaz M, Desseaux V, Marchis-Mouren G, Payan F, Forest E, and Santimone M. The mechanism of porcine pancreatic alpha-amylase. Kinetic evidence for two additional carbohydrate-binding sites. Eur J Biochem 241: 787-796, 1996. PMID: 8944767.
- Alpers DH. Digestion and Absorption of Carbohydrates and Proteins. In: Physiology of the Gastrointestinal Tract, edited by Johnson LR. New York, NY: Ravens Press, 1994, p. 1723-1749.
- Asanuma-Date K, Hirano Y, Le N, Sano K, Kawasaki N, Hashii N, Hiruta Y, Nakayama K, Umemura M, Ishikawa K, Sakagami H, and Ogawa H. Functional regulation of sugar assimilation by N-glycan-specific interaction of pancreatic alpha-amylase with glycoproteins of duodenal brush border membrane. J Biol Chem 287: 23104-23118, 2012. PMID: 22584580.
- Azzout-Marniche D, Chaumontet C, Piedcoq J, Khodorova N, Fromentin G, Tome D, Gaudichon C, and Even PC. High Pancreatic Amylase Expression Promotes Adiposity in Obesity-Prone Carbohydrate-Sensitive Rats. J Nutr 2019. PMID: 30753533.
- Bloor JH, Meisler MH, and Nielsen JT. Genetic determination of amylase synthesis in the mouse. J Biol Chem 256: 373-377, 1981. PMID: 6161122.
- Bompard-Gilles C, Rousseau P, Rouge P, and Payan F. Substrate mimicry in the active center of a mammalian alpha-amylase: structural analysis of an enzyme-inhibitor complex. Structure 4: 1441-1452, 1996. PMID: 8994970.
- Brannon PM. Adaptation of the exocrine pancreas to diet. Annu Rev Nutr 10: 85-105, 1990. PMID: 2200477.
- Brayer GD, Luo Y, and Withers SG. The structure of human pancreatic alpha-amylase at 1.8 A resolution and comparisons with related enzymes. Protein Sci 4: 1730-1742, 1995. PMID: 8528071.
- Buisson G, Duee E, Haser R, and Payan F. Three dimensional structure of porcine pancreatic alpha-amylase at 2.9 A resolution. Role of calcium in structure and activity. EMBO J 6: 3909-3916, 1987. PMID: 3502087.
- Cadegiani FA, and Silva OS. Acarbose promotes remission of both early and late dumping syndromes in post-bariatric patients. Diabetes Metab Syndr Obes 9: 443-446, 2016. PMID: 27994477.
- Ceska M, Birath K, and Brown B. A new and rapid method for the clinical determination of alpha-amylase activities in human serum and urine. Optimal conditions. Clin Chim Acta 26: 437-444, 1969. PMID: 5358545.
- Clavien PA, Burgan S, and Moossa AR. Serum enzymes and other laboratory tests in acute pancreatitis. Br J Surg 76: 1234-1243, 1989. PMID: 2691011.
- Criddle DN, Booth DM, Mukherjee R, McLaughlin E, Green GM, Sutton R, Petersen OH, and Reeve JR, Jr. Cholecystokinin-58 and cholecystokinin-8 exhibit similar actions on calcium signaling, zymogen secretion, and cell fate in murine pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol 297: G1085-1092, 2009. PMID: 19815626.
- D'Amico S, Gerday C, and Feller G. Structural similarities and evolutionary relationships in chloride-dependent alpha-amylases. Gene 253: 95-105, 2000. PMID:10925206.
- Date K, Satoh A, Iida K, and Ogawa H. Pancreatic alpha-Amylase Controls Glucose Assimilation by Duodenal Retrieval through N-Glycan-specific Binding, Endocytosis, and Degradation. J Biol Chem 290: 17439-17450, 2015. PMID: 26023238.
- Dhital S, Warren FJ, Butterworth PJ, Ellis PR, and Gidley MJ. Mechanisms of starch digestion by alpha-amylase-Structural basis for kinetic properties. Crit Rev Food Sci Nutr 57: 875-892, 2017. PMID: 25751598.
- Dranginis A, Morley M, Nesbitt M, Rosenblum BB, and Meisler MH. Independent regulation of nonallelic pancreatic amylase genes in diabetic mice. J Biol Chem 259: 12216-12219, 1984. PMID: 6207174.
- Erchinger F, Engjom T, Tjora E, Aksnes L, Dimcevskir G, and Gudbrandsen OA. Analysis of amylase in duodenal juice - Automated kinetic spectrophotometric analysis versus manual colorimetric endpoint assay. Pancreatology 2017. PMID: 28190684.
- Ferey-Roux G, Perrier J, Forest E, Marchis-Mouren G, Puigserver A, and Santimone M. The human pancreatic alpha-amylase isoforms: isolation, structural studies and kinetics of inhibition by acarbose. Biochim Biophys Acta 1388: 10-20, 1998. PMID: 9774702.
- Gilles C, Astier JP, Marchis-Mouren G, Cambillau C, and Payan F. Crystal structure of pig pancreatic alpha-amylase isoenzyme II, in complex with the carbohydrate inhibitor acarbose. Eur J Biochem 238: 561-569, 1996. PMID: 8681972.
- Giorgi D, Bernard JP, Lapointe R, and Dagorn JC. Regulation of amylase messenger RNA concentration in rat pancreas by food content. EMBO J 3: 1521-1524, 1984. PMID: 6204864.
- Gullo L, Lucrezio L, Migliori M, Bassi M, Nestico V, and Costa PL. Benign pancreatic hyperenzymemia or Gullo's syndrome. Adv Med Sci 53: 1-5, 2008. PMID:18650145.
- Gumucio DL, Wiebauer K, Caldwell RM, Samuelson LC, and Meisler MH. Concerted evolution of human amylase genes. Mol Cell Biol 8: 1197-1205, 1988. PMID: 2452973.
- Hiele M, Ghoos Y, Rutgeerts P, and Vantrappen G. Starch digestion in normal subjects and patients with pancreatic disease, using a 13CO2 breath test. Gastroenterology 96: 503-509, 1989. PMID: 2492013.
- Ismail OZ, and Bhayana V. Lipase or amylase for the diagnosis of acute pancreatitis? Clin Biochem 50: 1275-1280, 2017. PMID:28720341.
- Jung DH. Preparation and application of Procion Yellow starch for amylase assay. Clin Chim Acta 100: 7-11, 1980. PMID: 7351080.
- Keller SA, Rosenberg MP, Johnson TM, Howard G, and Meisler MH. Regulation of amylase gene expression in diabetic mice is mediated by a cis-acting upstream element close to the pancreas-specific enhancer. Genes Dev 4: 1316-1321, 1990. PMID: 1699843.
- Klein B, and Foreman JA. Amylolysis of a chromogenic substrate, Cibachron Blue F3GA-amylose: kinetics and mechanism. Clin Chem 26: 250-253, 1980. PMID: 6153298.
- Korc M, Owerbach D, Quinto C, and Rutter WJ. Pancreatic islet-acinar cell interaction: amylase messenger RNA levels ar determined by insulin. Science 213: 351-353, 1981. PMID: 6166044.
- Layer P, Zinsmeister AR, and DiMagno EP. Effects of decreasing intraluminal amylase activity on starch digestion and postprandial gastrointestinal function in humans. Gastroenterology 91: 41-48, 1986. PMID: 2423408.
- Mandel AL, Peyrot des Gachons C, Plank KL, Alarcon S, and Breslin PA. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One 5: e13352, 2010. PMID: 20967220.
- Matsushita H, Takenaka M, and Ogawa H. Porcine pancreatic alpha-amylase shows binding activity toward N-linked oligosaccharides of glycoproteins. J Biol Chem 277: 4680-4686, 2002. PMID: 11741899.
- Matthews EK, Petersen OH, and Williams JA. Analysis of tissue amylase output by an automated method. Anal Biochem 58: 155-160, 1974. PMID: 4825371.
- Maurus R, Begum A, Kuo HH, Racaza A, Numao S, Andersen C, Tams JW, Vind J, Overall CM, Withers SG, and Brayer GD. Structural and mechanistic studies of chloride induced activation of human pancreatic alpha-amylase. Protein Sci 14: 743-755, 2005. PMID:15722449.
- McInnes N, Smith A, Otto R, Vandermey J, Punthakee Z, Sherifali D, Balasubramanian K, Hall S, and Gerstein HC. Piloting a Remission Strategy in Type 2 Diabetes: Results of a Randomized Controlled Trial. J Clin Endocrinol Metab 2017. PMID: 28324049.
- Mehta DI, Wang HH, Akins RE, Wang L, and Proujansky R. Isolated pancreatic amylase deficiency: probable error in maturation. J Pediatr 136: 844-846, 2000. PMID: 10839889.
- Meisler MH, and Ting CN. The remarkable evolutionary history of the human amylase genes. Crit Rev Oral Biol Med 4: 503-509, 1993. PMID:7690604.
- Metz DC, Patto RJ, Mrozinski JE, Jr., Jensen RT, Turner RJ, and Gardner JD. Thapsigargin defines the roles of cellular calcium in secretagogue-stimulated enzyme secretion from pancreatic acini. J Biol Chem 267: 20620-20629, 1992. PMID: 1383205.
- Mifflin TE, Benjamin DC, and Bruns DE. Rapid quantitative, specific measurement of pancreatic amylase in serum with use of a monoclonal antibody. Clin Chem 31: 1283-1288, 1985. PMID: 3874721.
- Nakamura Y, Ogawa M, Nishide T, Emi M, Kosaki G, Himeno S, and Matsubara K. Sequences of cDNAs for human salivary and pancreatic alpha-amylases. In: GeneElsevier, 1984, p. 263-270.
- Orebaugh SL. Normal amylase levels in the presentation of acute pancreatitis. Am J Emerg Med 12: 21-24, 1994. PMID: 7506911.
- Owen DG, and Jordan CC. A modified amylase assay, using a fluorescent substrate, and its application to a study of the rat parotid gland in vitro. J Pharmacol Methods 6: 281-293, 1981. PMID: 6174829.
- Pasero L, Mazzei-Pierron Y, Abadie B, Chicheportiche Y, and Marchis-Mouren G. Complete amino acid sequence and location of the five disulfide bridges in porcine pancreatic alpha-amylase. Biochim Biophys Acta 869: 147-157, 1986. PMID: 3484639.
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, Carter NP, Lee C, and Stone AC. Diet and the evolution of human amylase gene copy number variation. Nat Genet 39: 1256-1260, 2007. PMID: 17828263.
- Pieper-Bigelow C, Strocchi A, and Levitt MD. Where does serum amylase come from and where does it go? Gastroenterol Clin North Am 19: 793-810, 1990. PMID: 1702756.
- Rinderknecht H. Pancreatic Secretory Enzymes. In: The Pancreas: Biology, Pathobiology, and Disease, edited by Go VLW. New York: Raven Press, 1993, p. 219-251.
- Rinderknecht H, Wilding P, and Haverback BJ. A new method for the determination of alpha-amylase. Experientia 23: 805, 1967. PMID: 6076299.
- Santimone M, Koukiekolo R, Moreau Y, Le Berre V, Rouge P, Marchis-Mouren G, and Desseaux V. Porcine pancreatic alpha-amylase inhibition by the kidney bean (Phaseolus vulgaris) inhibitor (alpha-AI1) and structural changes in the alpha-amylase inhibitor complex. Biochim Biophys Acta 1696: 181-190, 2004. PMID: 14871659.
- Santos JL, Saus E, Smalley SV, Cataldo LR, Alberti G, Parada J, Gratacos M, and Estivill X. Copy number polymorphism of the salivary amylase gene: implications in human nutrition research. J Nutrigenet Nutrigenomics 5: 117-131, 2012. PMID: 22965187.
- Schibler U, Pittet AC, Young RA, Hagenbuchle O, Tosi M, Gellman S, and Wellauer PK. The mouse alpha-amylase multigene family. Sequence organization of members expressed in the pancreas, salivary gland and liver. J Mol Biol 155: 247-266, 1982. PMID:6176715.
- Seigner C, Prodanov E, and Marchis-Mouren G. The determination of subsite binding energies of porcine pancreatic alpha-amylase by comparing hydrolytic activity towards substrates. Biochim Biophys Acta 913: 200-209, 1987. PMID: 3496119.
- Singla RK, Singh R, and Dubey AK. Important Aspects of Post-Prandial Antidiabetic Drug, Acarbose. Curr Top Med Chem 16: 2625-2633, 2016. PMID: 27086787.
- Sjolund K, Haggmark A, Ihse I, Skude G, Karnstrom U, and Wikander M. Selective deficiency of pancreatic amylase. Gut 32: 546-548, 1991. PMID: 1710200.
- Snook JT. Dietary regulation of pancreatic enzymes in the rat with emphasis on carbohydrate. Am J Physiol 221: 1383-1387, 1971. PMID: 5124283.
- Stiefel DJ, and Keller PJ. Preparation and some properties of human pancreatic amylase including a comparison with human parotid amylase. Biochim Biophys Acta 302: 345-361, 1973. PMID: 4699244.
- Sugino H. Comparative genomic analysis of the mouse and rat amylase multigene family. FEBS Lett 581: 355-360, 2007. PMID: 17223109.
- Svensson B, Fukuda K, Nielsen PK, and Bonsager BC. Proteinaceous alpha-amylase inhibitors. Biochim Biophys Acta 1696: 145-156, 2004. PMID: 14871655.
- Tietz NW, Burlina A, Gerhardt W, Junge W, Malfertheiner P, Murai T, Otte M, Stein W, Gerber M, Klein G, and et al. Multicenter evaluation of a specific pancreatic isoamylase assay based on a double monoclonal-antibody technique. Clin Chem 34: 2096-2102, 1988. PMID: 3048784.
- Tosi M, Bovey R, Astolfi S, Bodary S, Meisler M, and Wellauer PK. Multiple non-allelic genes encoding pancreatic alpha-amylase of mouse are expressed in a strain-specific fashion. EMBO J 3: 2809-2816, 1984. PMID:6098446.
- Tsai A, Cowan MR, Johnson DG, and Brannon PM. Regulation of pancreatic amylase and lipase gene expression by diet and insulin in diabetic rats. Am J Physiol 267: G575-583, 1994. PMID: 7524347.
- Tsujita T. Persimmon-Tannin, an alpha-Amylase Inhibitor, Retards Carbohydrate Absorption in Rats. J Nutr Sci Vitaminol (Tokyo) 62: 192-197, 2016. PMID:27465726.
- Vasella A, Davies GJ, and Bohm M. Glycosidase mechanisms. Curr Opin Chem Biol 6: 619-629, 2002. PMID:12413546.
- Vissers RJ, Abu-Laban RB, and McHugh DF. Amylase and lipase in the emergency department evaluation of acute pancreatitis. J Emerg Med 17: 1027-1037, 1999. PMID: 10595892.
- Visvanathan R, Jayathilake C, and Liyanage R. A simple microplate-based method for the determination of alpha-amylase activity using the glucose assay kit (GOD method). Food Chem 211: 853-859, 2016. PMID: 27283705.
- Whitcomb DC, and Lowe ME. Human pancreatic digestive enzymes. Dig Dis Sci 52: 1-17, 2007. PMID: 17205399.
- Williams JA, Korc M, and Dormer RL. Action of secretagogues on a new preparation of functionally intact, isolated pancreatic acini. Am J Physiol 235: 517-524, 1978. PMID: 215042.
- Wise RJ, Karn RC, Larsen SH, Hodes ME, Gardell SJ, and Rutter WJ. A complementary DNA sequence that predicts a human pancreatic amylase primary structure consistent with the electrophoretic mobility of the common isozyme, Amy2 A. Mol Biol Med 2: 307-322, 1984. PMID: 6336237.
- Yokouchi H, Horii A, Emi M, Tomita N, Doi S, Ogawa M, Mori T, and Matsubara K. Cloning and characterization of a third type of human alpha-amylase gene, AMY2B. Gene 90: 281-286, 1990. PMID: 2401405.
- Zhang X, Caner S, Kwan E, Li C, Brayer GD, and Withers SG. Evaluation of the Significance of Starch Surface Binding Sites on Human Pancreatic alpha-Amylase. Biochemistry 55: 6000-6009, 2016. PMID:27756128.